A Multi-Resolution Strategy to Study Mechanisms of αβ-tubulin dimer Biogenesis,
Microtubule polymerization Regulators, and Mitotic Motor proteins:
Microtubules are dynamic polymers that are critical for many physical transformations that eukaryotic cells undertake in order to undergo cell division, development or crawl and move. Microtubules assemble from basic building blocks, called alpha beta (αβ) tubulin dimers. Microtubule polarized polymerization and dynamic instability stem from properties of their building blocks, αβ-tubulin dimers, which polymerize head-to-tail to form protofilaments, using the energy of Guanosine 3,5 Triphosphate (GTP) binding and hydrolysis. These tubulin protofilaments associate laterally enclosing a tube-like structure — a microtubule. Tubulin polymerization activates GTP hydrolysis only within the microtubule wall and leads to stochastic events called catastrophes, in which protofilaments peel and curl to disassemble the microtubule structure. The dynamics of tubulin heterodimer assembly and disassembly into microtubules occurs at their ends, and are highly regulated by conserved types of proteins that are found in all organisms. These proteins, in effect, act as MT “Polymerases” and “Depolymerases” accelerating the slow processes of polymerization and depolymerization that tubulin dimers can undergo alone. The laboratory is focused on deciphering the mechanisms of these microtubule regulatory proteins and how they cooperate to regulate microtubule dynamics in highly synchronized cellular phenomena like cell division and development. 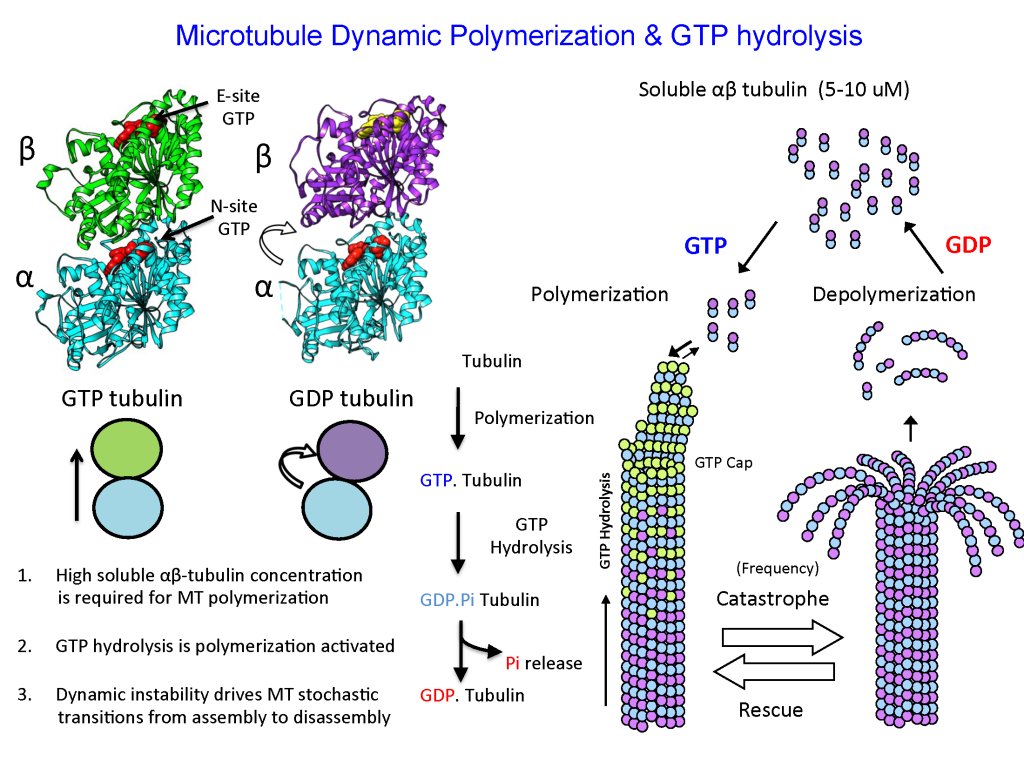
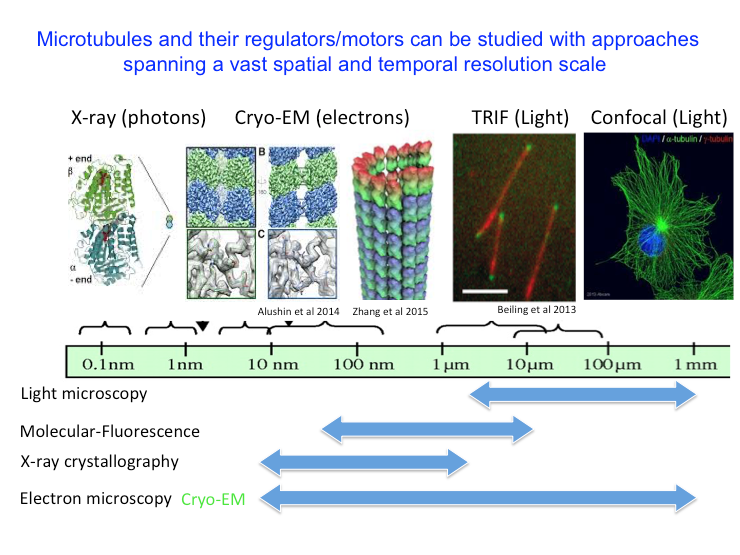
Microtubule structure and organization can be studied by multiple approaches that span a large resolution scale from micro-meters to the nanometers. Light microscopy can visualize microtubules in cells. High resolution total internal reflection fluorescence (TRIF) microscopy can be used to visualize the dynamics of microtubules in vitro and localization of regulatory proteins at their ends. Cryo-electron microscopy (Cryo-EM) can be used to visualize microtubule and tubulin structure. X-ray crystallography can be used to visualize the atomic structure of αβ-tubulins in complex with proteins. Our laboratory combines biochemical reconstitution, structural biology approaches such as cryo-electron microscopy and X-ray crystallography, with high-resolution single molecule total internal reflection fluorescence (TIRF) microscopy to study the mechanisms of Microtubule Polymerases, polymerization regulators, and mitotic motor proteins across multiple levels of spatial and temporal resolution.
Tubulin Biogenesis forms soluble αβ-tubulin pools and impacts Microtubule Function
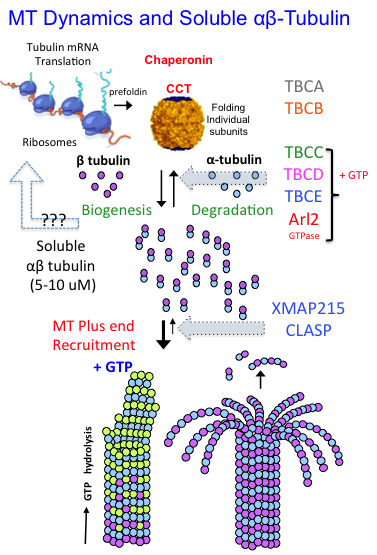
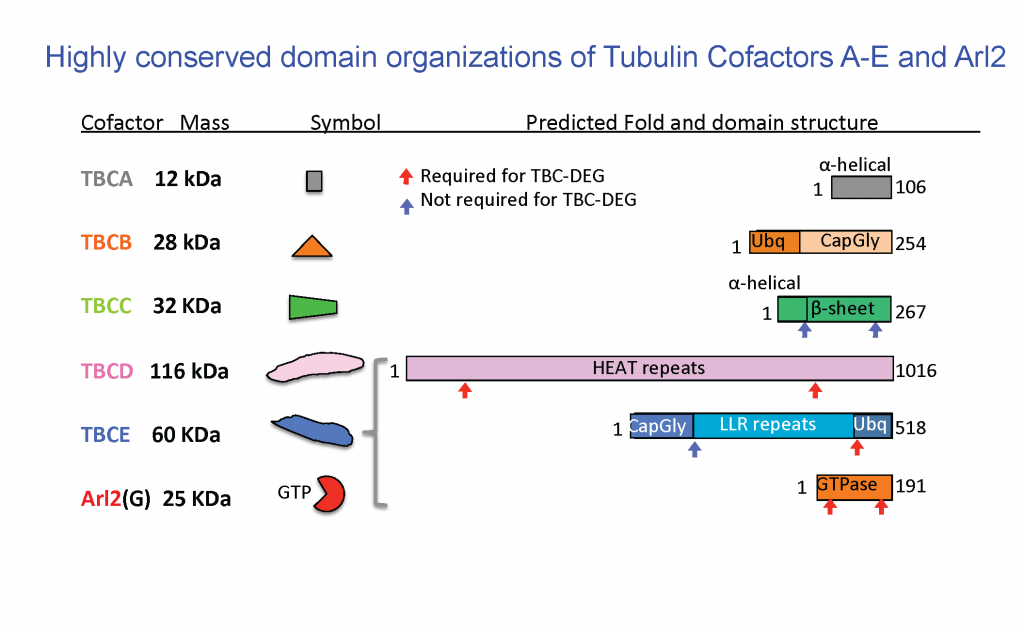
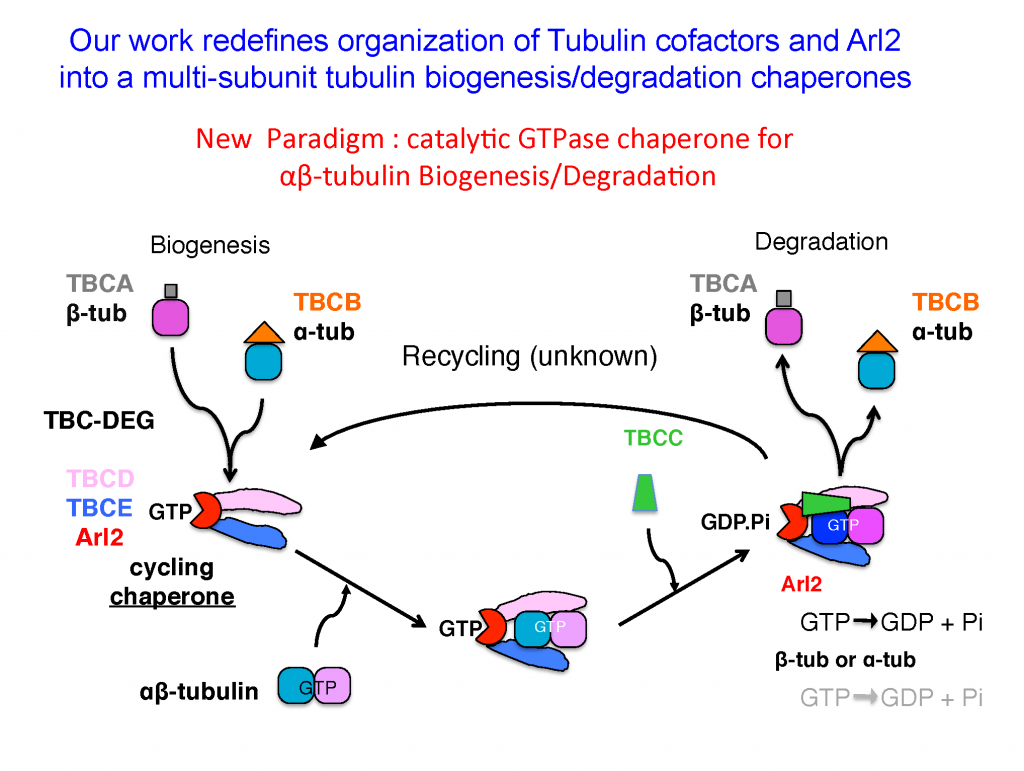
Eukaryotic cells build concentrated pools of αβ-tubulin dimers in the cytoplasm through high levels of translation from stable α and β-tubulin mRNAs. The assembly of αβ-tubulin from the newly folded α-tubulins and β-tubulins is mediated by five tubulin cofactors and Arl2, which is an Arf-like GTPase. Four of these molecules form a multi-subunit assembly which mediates GTP dependent catalytic assembly of αβ-tubulin through a process termed αβ-tubulin biogenesis. Our laboratory has demonstrated the shared functions of four subunits as a multi-subunit catalytic chaperone that forms a platform for αβ-tubulin biogenesis. The regulation of soluble αβ-tubulins by this biogenesis assembly process assembles the concentrated pools of αβ-tubulin in the cytoplasm and directly impacts microtubule dynamic functions across eukaryotes.
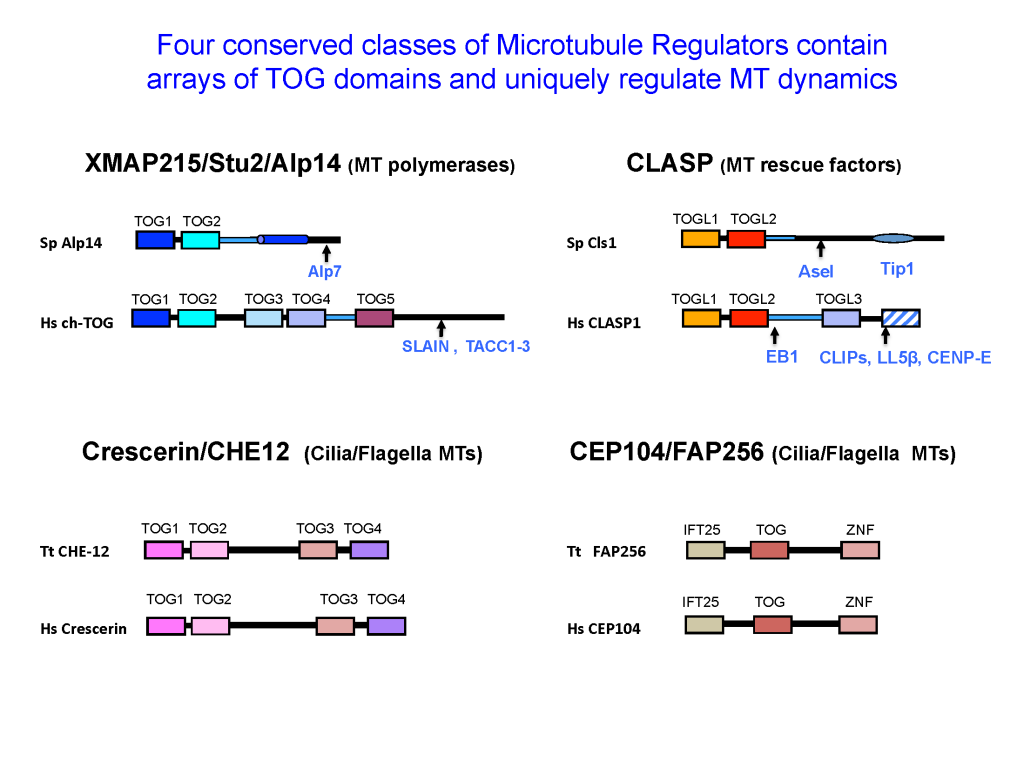
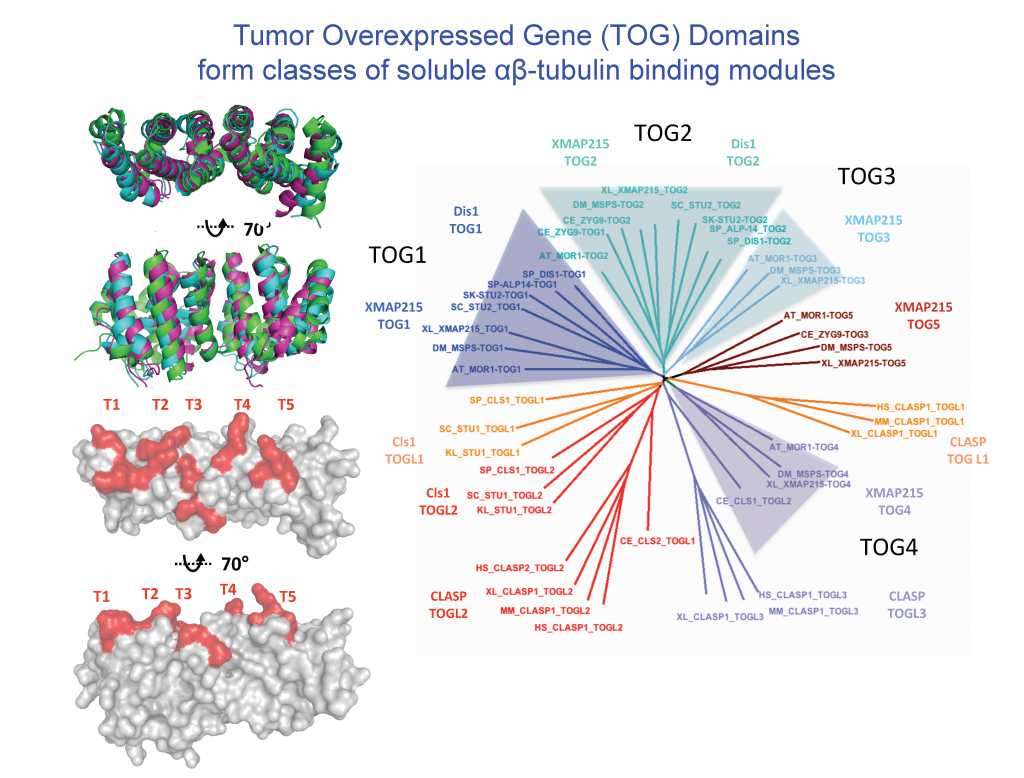
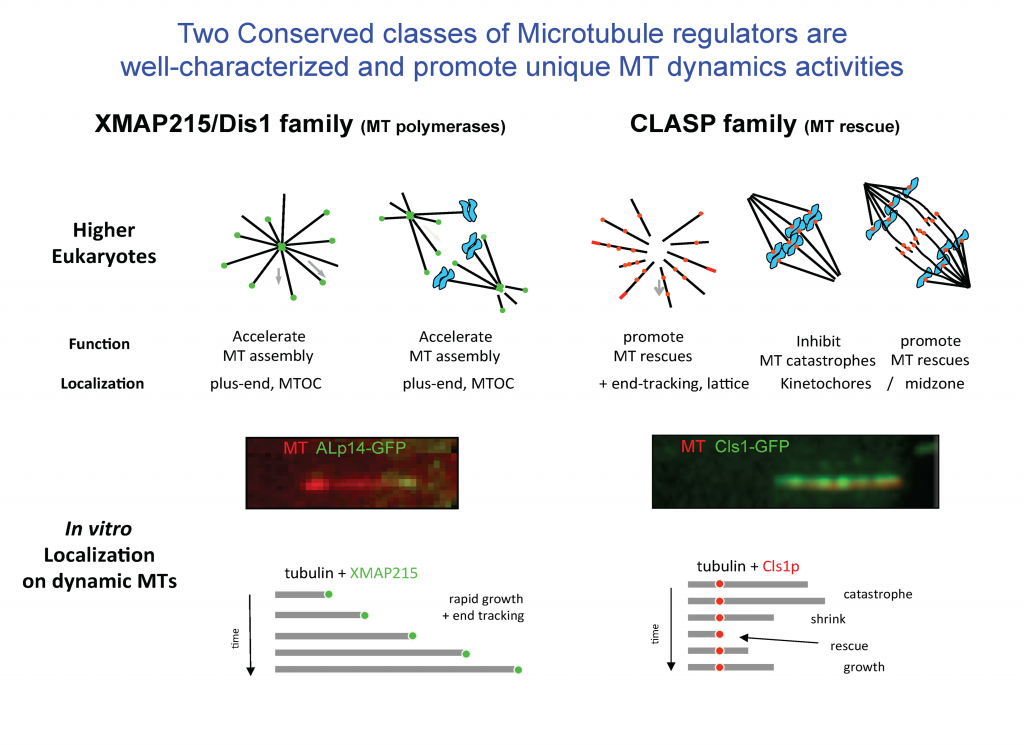
Four classes of Regulators recruit αβ-tubulins via TOG domains to modulate microtubule dynamics
The dynamics polymerization and depolymerization of soluble αβ-tubulins into microtubules occurs at their ends. Although αβ-tubulin polymerization into microtubule ends is spontaneous in vitro, it is not spontaneous in living cells. Microtubule polymerization requires highly regulated by conserved regulators that are found in all eukaryotic organisms. These proteins, in effect, act as microtubule “Polymerases” to accelerate the slow processes of polymerization by recruiting αβ-tubulins to microtubule ends. These proteins share the presence of conserved arrays of Tumor Overexpressed Gene (TOG) domains. TOG domains recruit αβ-tubulins to microtubule ends and regulate microtubule polymerization. The laboratory is focused on deciphering the mechanisms of these microtubule regulatory proteins and how they cooperate to regulate microtubule dynamics in highly synchronized cellular phenomena like cell division and development
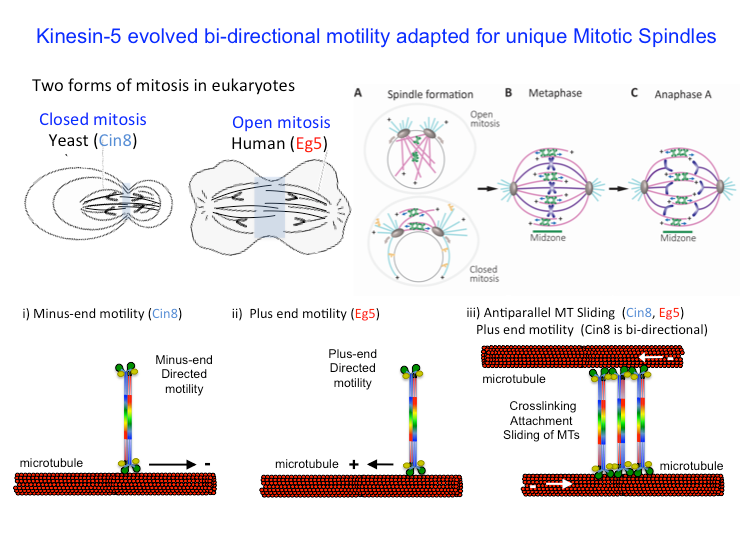
The Mitotic Kinesin-5 Motors are Bipolar Tetramers which mediate microtubule sliding
We are interested in understanding the mechanisms of kinesin-5 motors. These motors are unique tetrameric assemblies that mediate the sliding apart of microtubules leading to the bipolar organization of the mitotic spindles. Our laboratory has determined the structures of a major section of the bipolar mini-filament which lead to a unique model for how the the sliding apart of antiparallel microtubules. We also study the unique yeast Kinesin-5 motors which are mediate a unique reversal of direction towards microtubule minus ends as tetramers and reverse direction toward microtubule plus-ends as they assemble into higher oligomers leading to microtubule sliding.