2020
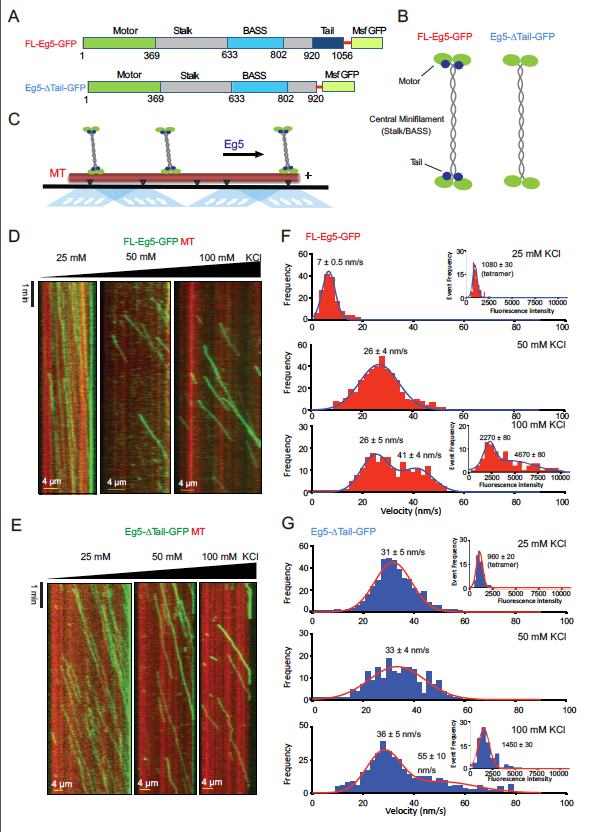
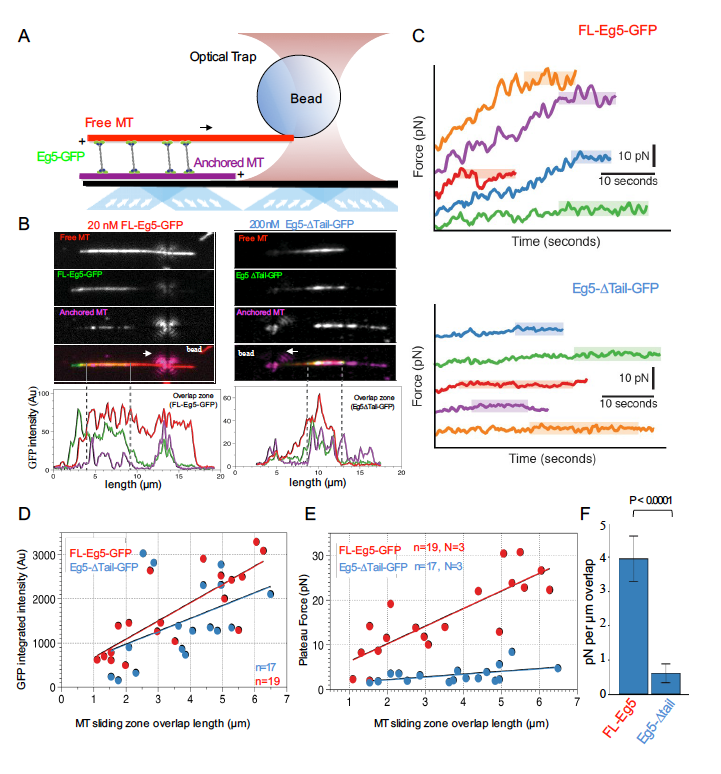
The kinesin-5 tail domain directly modulates the mechanochemical cycle of the
motor domain for anti-parallel microtubule sliding
Published in eLife
News coverage: UC-Davis News
Kinesin-5 motors organize mitotic spindles by sliding apart microtubules. They are homotetramers with dimeric motor and tail domains at both ends of a bipolar minifilament. Here, we describe a regulatory mechanism involving direct binding between tail and motor domains and its fundamental role in microtubule sliding. Kinesin-5 tails decrease microtubule-stimulated ATP-hydrolysis by specifically engaging motor domains in the nucleotide-free or ADP states. Cryo-EM reveals that tail binding stabilizes an open motor domain ATP-active site. Full-length motors undergo slow motility and cluster together along microtubules, while tail-deleted motors exhibit rapid motility without clustering. The tail is critical for motors to zipper together two microtubules by generating substantial sliding forces. The tail is essential for mitotic spindle localization, which becomes severely reduced in tail-deleted motors. Our studies suggest a revised microtubule-sliding model, in which kinesin-5 tails stabilize motor domains in the microtubule-bound state by slowing ATP-binding, resulting in high-force production at both homotetramer ends.
2019
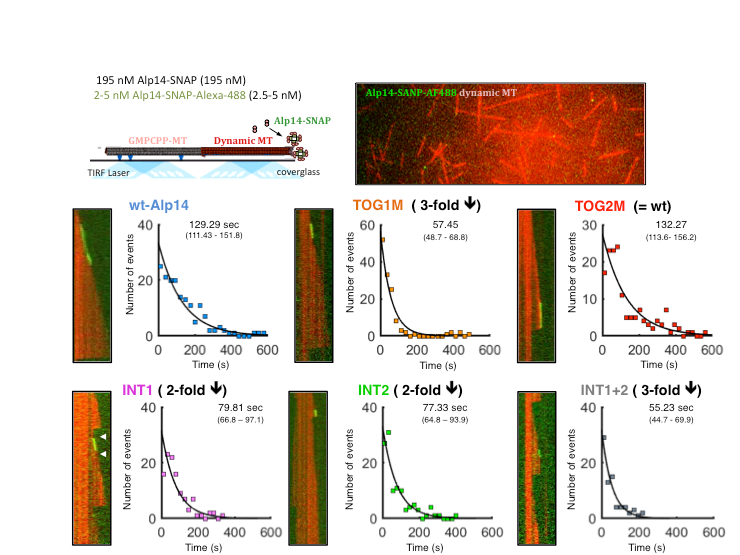
Microtubule polymerase and processive plus-end tracking functions originate from
distinct features within TOG domain arrays
Published in Molecular Biology of the Cell
Microtubule polymerases accelerates tubulin polymerization while processively tracking microtubule (MT) plus ends via tumor overexpressed gene (TOG) domain arrays. We describe a relationship between MT plus-end tracking and polymerase functions revealing their distinct origin within TOG arrays using structural based model-based mutants, microtubule dynamic assays, single molecule fluorescence and studies of the mutants within fission yeast cells. We study Alp14 mutants designed based on structural models, with defects in either tubulin recruitment or self-organization. Using in vivo live imaging in fission yeast and in vitro MT dynamics assays, we show that tubulins recruited by TOG1 and TOG2 serve concerted, yet distinct, roles in MT plus-end tracking and polymerase functions. TOG1 is critical for processive plus-end tracking, whereas TOG2 is critical for accelerating tubulin polymerization. Inactivating interfaces that stabilize square complexes lead to defects in both processive MT plus-end tracking and polymerase. Our studies suggest that a dynamic cycle between square and unfurled TOG array states gives rise to processive polymerase activity at MT plus ends.
2018
Structural Basis for the Recruitment and Polymerization of αß-tubulins by proteins with Tumor Overexpressed Gene (TOG) domains: Structural Basis of a Microtubule Polymerase
Published in eLife
News coverage: Phys.org, UC-Davis Biology
The polymerization of αß-tubulins at microtubule plus-ends is accelerated by conserved proteins with arrays of Tumor Overexpressed Gene (TOG) domains. These proteins form microtubule polymerases which accelerate microtubule polymerization by recruiting αß-tubulins to microtubules while processively tracking their plus-ends. These proteins are essential regulators and their defects are identified in liver and colon cancers and lead to defects in cell division due to poor microtubule dynamics in a variety of organisms. Although it is known that arrays of TOG domains recruit αß-tubulins to microtubule ends, their mechanisms as polymerases remain poorly understood.
Our group has determined the biochemical and high resolution structural states for TOG arrays in complex with αβ-tubulins in the recruitment and polymerization states. These studies in combination with molecular modeling, mutagenesis studies leads to a brand new polarized unfurling model for their shared mechanisms as microtubule polymerases that accelerate αβ-tubulin polymerization at microtubule ends. The model is described in stills and movie shown below. In this model, dimeric TOG1-TOG2 arrays recruit four αβ-tubulins and form TOG square assemblies, which inhibit spontaneous αβ-tubulin polymerization, while they diffuse along microtubules s via positively charged regions. When these assemblies reach MT plus-ends, they may dock onto microtubule plus-ends via αβ-tubulins bound to either TOG1 or TOG2. Molecular modeling shows that docking via TOG1:αβ-tubulin, and not via TOG2:αβ-tubulin, leads a steric clash with the microtubule wall, promoting the unfurling of the TOG square state to form corner-like TOG1-TOG2 arrays:αβ-tubulin complexes. These corner-like TOG1-TOG2 arrays transition to polymerizing their αβ-tubulins into protofilaments, as observed in our second structure, through hinge-like rotation around the anchored TOG1:αβ-tubulin and mediated by Brownian motion. TOG2 rapidly dissociates from the tip of this new polymer followed by protofilament straightening and this cycle repeats. Thus, TOG2 is critical for accelerating αβ-tubulin polymerization while TOG1:αβ-tubulin anchors the assembly onto the MT plus-end. This model predicts that: 1) in order to persist in tracking MT-plus-ends, TOG square assemblies must reform to recruit new αβ-tubulins. 2) TOG1 and TOG2 serve different, yet complementary, roles in microtubule polymerase and plus-end tracking.
2015
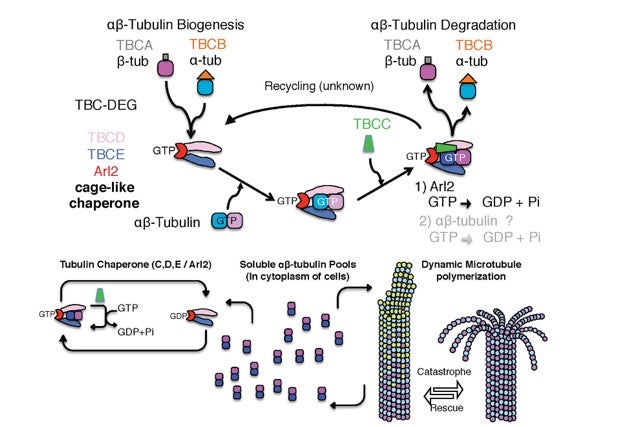
Tubulin cofactors and Arl2 form a cage-like chaperone that catalyzes αβ-tubulin assembly through GTP hydrolysis: Reconstitution of new molecular machine
Published in eLife
The building block of microtubules is composed of two proteins called α-tubulin and ß-tubulin which are assembled into a single polarized αß-tubulin subunit. Microtubules are very dynamic structures that can rapidly change length as individual αβ-tubulin units are either added or removed to their growing or shrinking ends. The assembly of aß-tubulin has for long remained a mystery despite extensive genetic and cell biology studies. Several proteins known as tubulin cofactors and an enzyme called Arl2 help to build a vast pool of αβ-tubulin that are able attach to the microtubules. It has remained unknown how this collection of proteins drives αβ-tubulin biogenesis and degradation inside cells. And the roles of the conserved tubulin cofactors and Arl2 remain unknown.
Our group used a combination of biochemical reconstitution of all tubulin cofactors and Arl2 GTPase in combination with negative stain electron microscopy and biochemical techniques to study how the tubulin cofactors and Arl2 are organized, and their role in the assembly of aß-tubulin and polymerization of dynamic microtubules inside living yeast cells. Our biochemical reconstitution has revealed that Arl2 and two tubulin cofactors TBCD and TBCE associate with each other to form a stable ‘complex’ that has a cage-like structure. A molecule of αβ-tubulin binds to the complex, followed by another cofactor called TBCC. The binding of αβ-tubulin and TBCC activates the enzyme activity of Arl2, which releases the energy needed to alter the shape of the αβ-tubulin in a larger ternary complex. We also found that inactivating Arl2 GTP hydrolysis inhibits the dissociation of this ternary complex and introducing this mutant form of Arl2 that lacked enzyme activity inside yeast cells had problems forming microtubules characterized by poor polymerization due to pausing and defects in cell division.
Together, our findings show that the tubulin cofactors and Arl2 form a complex that regulates the assembly and maintenance of αβ-tubulin. The next challenges are : 1) to understand how this regulation occurs by determining the high resolution structures of these assemblies and 2) to determine how this regulation influences the way that microtubules grow and shrink inside cells.
2014
Structural Basis for the Assembly of the Mitotic Motor Kinesin-5 into Bipolar Tetramers
Published in eLife
During cell division, an organized and symmetric intracellular apparatus, named the mitotic spindle, composed of microtubules (and associated molecular machines), coordinate the accurate alignment and equal distribution of newly duplicated chromosomes into two new cells. When the organization of mitotic spindles becomes mis-regulated, a variety of human disease conditions can result, including early onset cancers and human birth defects. Our laboratory studies the molecular mechanisms of microtubule dynamics and organization during mitotic cell division.
Molecular motors in living cells bind and generate motile forces through “step-by-step walking” along microtubules. These motor activities move chromosomes and mechanically orchestrate events during mitotic cell division. We are focused on an evolutionarily well-conserved molecular motor class, Kinesin-5, which are fundamentally responsible for elongating mitotic spindles to form two new cells. The organization of mitotic motor kinesin-5 is unique from all other molecular motors in that they form elongated bipolar molecular assemblies, composed of four subunits, with two sets of active motile motors pointed towards opposite ends of a rigid central rod. The central rod is critical for transmitting forces between the motile ends. The “dual-headed” arrangement of kinesin-5 is critical for its function as it preferentially binds microtubules emanating from opposite poles of the mitotic spindle and mediates their anti-parallel sliding near the end of the cell division cycle.
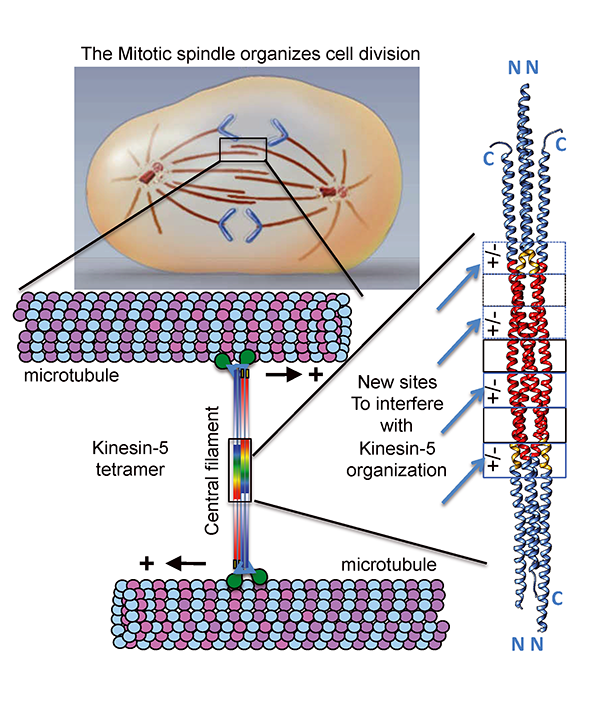 Our laboratory made a major breakthrough, in collaboration with Jonathan Scholey’s group at UC-Davis, in determining the atomic three-dimensional structure four-subunit kinesin-5 central rod region, which specifies the mitotic kinesin-5 organization into “dual headed” bipolar molecular machines. We discovered a precise three-dimensional organization where four kinesin-5 subunits form bipolar assemblies. We observed an alternating pattern of seven symmetrically organized interface elements composed of uniquely interacting amino acids. Four out of seven of these interface elements are ionic in nature and form exposed “molecular pockets” that are accessible to small chemical molecules from the solution. Mutations of the ionic interacting amino acids severely disrupt the kinesin-5 four-subunit organization, leading misassembled kinesin-5 molecules. The kinesin-5 structure suggests a novel strategy in targeting kinesin-5 inactivation that is conceptually more effective in controlling mitotic spindle elongation than the previous strategies as it is specifically focused on disrupting kinesin-5 assembly by disassembling its central bipolar rod filament, the main region that transmits force between the two kinesin-5 motile ends.
Our laboratory made a major breakthrough, in collaboration with Jonathan Scholey’s group at UC-Davis, in determining the atomic three-dimensional structure four-subunit kinesin-5 central rod region, which specifies the mitotic kinesin-5 organization into “dual headed” bipolar molecular machines. We discovered a precise three-dimensional organization where four kinesin-5 subunits form bipolar assemblies. We observed an alternating pattern of seven symmetrically organized interface elements composed of uniquely interacting amino acids. Four out of seven of these interface elements are ionic in nature and form exposed “molecular pockets” that are accessible to small chemical molecules from the solution. Mutations of the ionic interacting amino acids severely disrupt the kinesin-5 four-subunit organization, leading misassembled kinesin-5 molecules. The kinesin-5 structure suggests a novel strategy in targeting kinesin-5 inactivation that is conceptually more effective in controlling mitotic spindle elongation than the previous strategies as it is specifically focused on disrupting kinesin-5 assembly by disassembling its central bipolar rod filament, the main region that transmits force between the two kinesin-5 motile ends.