We utilize a combination of the approaches, listed below, to span the spatial and temporal resolution scales to link structure with function. These allow for the isolation of macromolecular assemblies with biological activity. Using these approaches, we can purify, engineer and then visualize the process in which a protein acts as a machine and catch them at various steps of biological activity. We then develop structural hypotheses and introduce mutants to determine their activity and link structure with basic functional mechanism.
Expression and Engineering of
Recombinant Proteins and Multi-protein Complexes
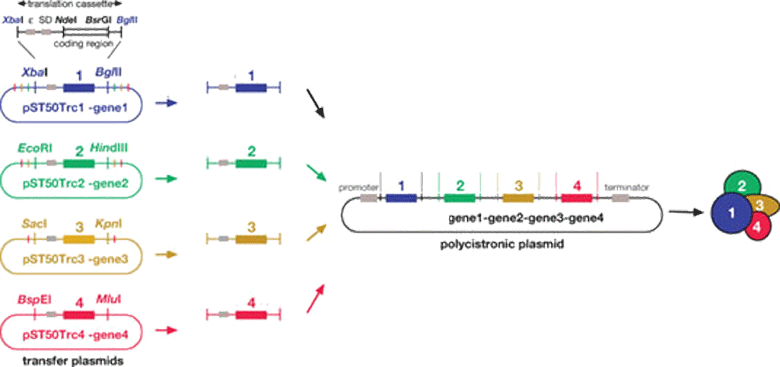
We express recombinant proteins in bacterial (Escherichia coli) and insect (Spodoptera frugiperda) and Tetrahymena (Tetrahymena thermophila) cell expression systems. We can isolate multi-protein complexes using poly-cistronic expression in these systems. Molecular biology approaches are used to design recombinant proteins based on their sequence conservation to isolate different domains for functional and structural analyses. Recombinant proteins or protein complexes are purified using a variety of chromatography techniques including affinity, ion exchange and size exclusion chromatography
Biophysical and Biochemical Mass and affinity Analyses of Protein Complexes
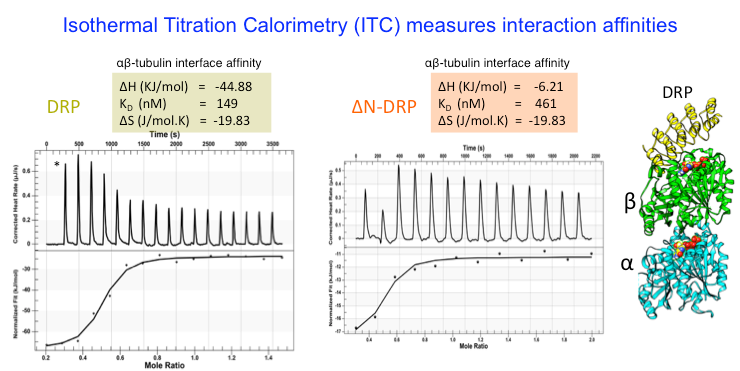
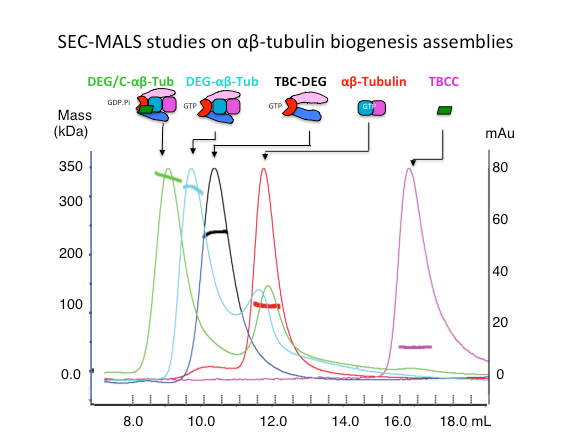
We analyze the biochemical mass, oligomerization, using Size Exclusion Chromatography (SEC), Size exclusion chromatography and Multi-Angle Light Scattering (SEC-MALS). We measure protein binding affinities using isothermal titration calorimetry (ITC) .
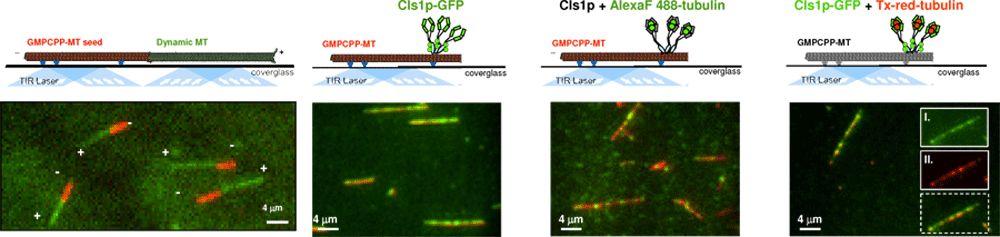
High Resolution Total Internal Fluorescence (TIRF) Microscopy Studies
to reconstitute activities of Microtubule Dynamics with their Regulators
The mechanism of multi-protein complexes in regulating MT dynamics can be studied using high resolution light microscopy approach, termed total internal reflection fluorescence microscopy, where a thin 50 nm slab of solution near the surface of a glass is illuminated through the “total reflection” of a fluorescent laser light.
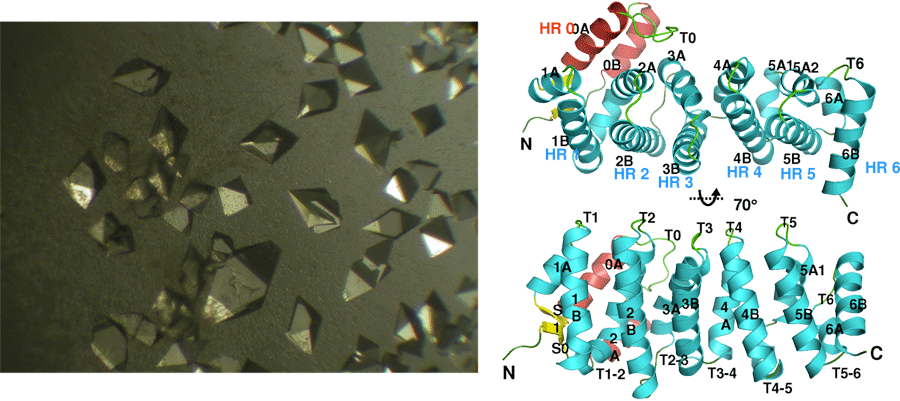
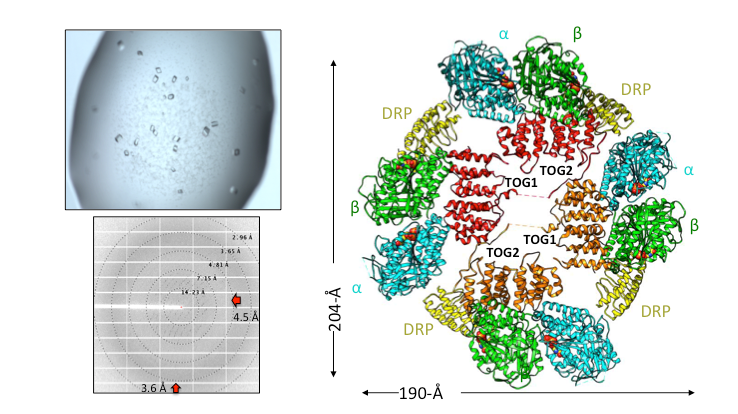
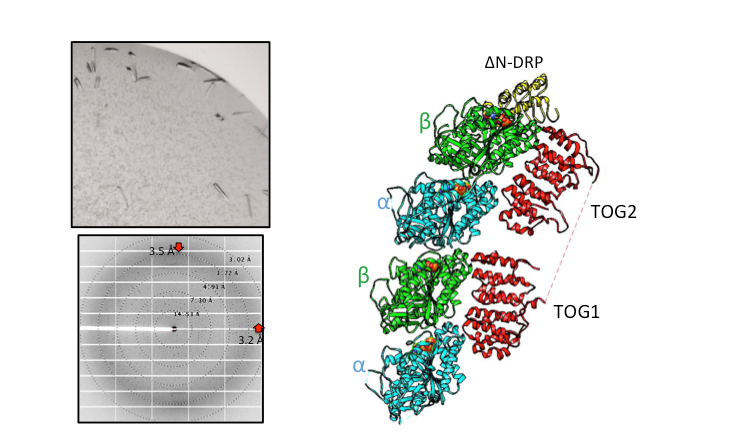
Structural Studies of Multi-subunit Protein complexes using X-ray Crystallography
The structures of individual recombinant proteins or protein domains can be determined using x-ray crystallography. Highly purified recombinant proteins form crystals when subjected to dehydrating and precipitating chemical environments. When these crystals are subjected to intense narrow x-ray beams at national radiation synchrotron sources, they generate diffraction patterns, which can then be solved using a variety of computational approaches to determine the molecular architecture of protein in the crystals.
Cryo-electron Microscopy (Cryo-EM) and single particle image analysis
to determine Structures of Multi-protein Complexes
The structures of multi-protein complexes in different states can be studied using molecular electron microscopy of negatively stained samples or cryo preserved in vitreous ice. Using computational approaches, medium or low-resolution three-dimensional reconstructions can be determined for protein complexes. High resolution structural studies can be performed using cryo-electron microscopy Cryo-EM. Our laboratory has access to the BIO-EM facility which includes a FEI Glacios microscope equipped with an autoloader and K3 direct electron detector, and using monthly data collection time as a part of the Bay Area Cryo-EM consortium (BACEM) using FEI titan Krios microscopes located at the Universities of California San Francisco and Berkeley. These high resolution structures allow De Novo model building or docking of high-resolution structures of component proteins.